Imaging software targets scientific applications
Andrew Wilson, Editor
Large and complex image data found in biology, medicine and environmental sciences demand specialized software to perform such tasks as cell analysis, 3D image reconstruction and multi-dimensional data visualization. To perform such tasks, a number of companies offer both specialized software to perform single specialized operations while others offer broad multi-function libraries.
Just as software packages used in machine vision use specific classes of algorithms to identify and extract image data, so do those that target specific scientific and medical applications such as the automatic identification of microscopy images. Like their machine vision counterparts, these software packages are designed to allow developers to configure automated systems that can analyze and classify multiple images at high data rates.
To accomplish this task, such systems are required to first identify relevant regions of interest (ROIs) within an image, extract image features and classify the images based on the extracted data. To highlight regions within such images, numerous techniques such as thresholding, edge detection, watershed-based algorithms, model-based segmentation and contouring methods can be used.
Often, these algorithms are tailored to detect specific features with an object – such as cells - while others must be tailored to accommodate the feature to be detected. To speed the analysis of identifying these regions of interest in microscope-based applications, captured images can be tiled and multiple tiles can be distributed between multi-core processors to speed image analysis.
Once such features are extracted, these features must be automatically classified. To perform this task, different types of classifiers can be trained with known data sets and tested against sample data to determine their effectiveness. Just as software developed for machine vision systems uses a number of different types of classifiers to perform this task, so too does the software developed for automated microscopy analysis. These classifiers include those based on nearest neighbor, boosting, random forest and support vector machines (SVMs).
Plug-ins and modules
Today, there are many commercially available and open source software packages that provide researchers with the ability to perform such image analysis and classification. Because image analysis applications in biology, medicine and environmental sciences differ widely, the algorithms used for image analysis and classification must often be tailored for these specific applications. Understanding this, many commercially available packages such as Image-Pro Plus from Media Cybernetics (Rockville, MD, USA;www.mediacy.com) and MetaMorph microscopy and automation software from Molecular Devices (Sunnyvale, CA, USA; www.moleculardevices.com) are modular packages that offer different plug-ins and modules designed to target specific microscopy applications.
Media Cybernetics's Image-Pro Plus software, for example, offers numerous image processing algorithms to analyze images from both manual and automated microscopy systems. These include functions to capture, analyze, measure and classify images, track moving objects and perform measurements on 3D rendered images. As well, the company offers a number of more specialized software packages such as the company's Gel-Pro Analyzer that allows information to be extracted from electrophoretic gels, blots and colonies. To tailor Image-Pro for any specific application, the package's Software Development Kit (SDK) allows systems integrators to develop custom plug-ins and add them into existing software menus and toolbars. In this way, Image-Pro Plus can be tailored to suit the requirements of specific vertical markets.
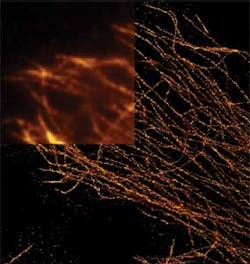
Like ImagePro, Molecular Devices' MetaMorph software also performs image acquisition, processing and analysis functions from microscopy-based systems. As well, the software can be used to control many of the peripherals required in automated microscopy systems such as microscopes, filter wheels, shutters, cameras, piezo-electric focus devices and motorized stages. The software also includes software applications modules tailored to perform such functions as cell counting, image deconvolution, motion analysis, particle tracking and volume measurements (Figure 1).
While commercially available packages such as ImagePro and MetaMorph are specifically tailored to these scientific applications, a number of more general purpose image processing packages are available both commercially and in the public domain. These include the commercially available Image Processing Toolbox from MathWorks (Natick, MA, USA;www.mathworks.com), an image processing package that provides a set of standard algorithms, functions, and applications for image processing, analysis, visualization, and algorithm development. Using the software, developers can analyze, segment and enhance images and perform functions such as geometric transformations and image registration.
Tailored software
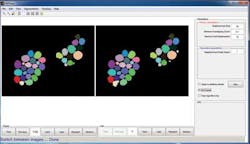
While the Image Processing Toolbox is a general purpose image processing package, Mike West, Quanli Wang and Lingchong You at Duke University (Durham NC, USA;www.duke.edu) have used MathWorks' MATLAB and the Toolbox to create an open-source graphical user interface to implement novel methods for automating cell segmentation and reconstruction. Dubbed CellTracer, the software is designed for biologists without training in image processing or programing to extract single cell information from microscopy images (Figure 2). Freely available at http://bit.ly/1dNk9n9, the software requires MATLAB and the MATLAB image processing tool box to be installed on a host computer.
Like CellTracer, CellProfiler is free open-source software designed to enable biologists to quantitatively measure phenotypes from thousands of images automatically. Originally developed by Anne Carpenter and Thouis Jones of the Broad Institute (Cambridge, MA, USA;www.broadinstitute.org) using MATLAB, the latest incarnation of the software, CellProfiler 2.0 is written in Python and is designed for quantifying data from biological images, particularly in high-throughput experiments.
Functions of the software include the measurement of size, shape, intensity, and texture of every cell (or other object) in an image. In operation, a graphical user interface (GUI) allows developers to construct an image analysis pipeline – a sequential series of modules that perform an image processing function such as illumination correction, object identification (segmentation) and object measurement.
A companion software package – also written in Python - CellProfiler Analyst allows large, high-dimensional image-derived data to be analyzed and includes machine learning tools for identifying complex and subtle phenotypes. The compiled software is freely available for Macintosh, PC, and UNIX operating systems atwww.cellprofiler.org.
While CellTracer and CellProfiler allow features of images to be analyzed and tabulated, more specialized imaging software is also available that allows cell movements and divisions to be individually visualized and tracked. In this way, researchers can better understand how a tissue will change over time. GoFigure2, created by the Megason Lab in the Department of Systems Biology at Harvard Medical School (Boston, MA, USA;www.hms.harvard.edu), was created specifically for this purpose. Available under a BSD open source license and written in C++, the software is designed for the visualization, interaction, and analysis of large image sets from confocal microscopes.
Open-source code
Just as developers have used commercially available image processing software to develop software packages for specialized scientific applications, general purpose open source software packages have been used in a similar manner. One of these, ImageJ, written by Wayne Rasband of the Research Services Branch at the National Institute of Mental Health (Bethesda, MD, USA;www.nimh.nih.gov) is a public domain Java image processing program that can be downloaded at http://imagej.nih.gov/ij.
Running under Windows, Mac OS, and Linux, the software can be used to calculate area and pixel value statistics, measure distances and angles, create density histograms and line profile plots and supports image processing functions such as contrast manipulation, sharpening, smoothing, edge detection and median filtering. Since the software is multithreaded, time-consuming operations can be performed in parallel with other operations.
Since ImageJ supports plug-ins, many developers have written software components that add specific features for specialized applications. One such plug -in dubbed NeuronJ has been developed by Dr. Erik Meijering Associate Professor with the Biomedical Imaging Group Rotterdam at the University Medical Center Rotterdam (Rotterdam, The Netherlands;www.bigr.nl). Designed to measure the lengths of neurites, the semi-automatic tracing software has been used to study the development of the nervous system.
Like NeuronJ, NeuriteTracer developed by Madeline Pool and her colleagues at the Fournier Lab of McGill University's Neurological Institute (Montreal, Quebec, Canada; http://fournierlab.mcgill.ca) is also designed to replace the labor-intensive tracing of neurites.
To automate these measurements, NeuriteTracer, another freely available plug-in for ImageJ, can be used to analyze fluorescence microscopy images of neurites and nuclei of neurons, counting neuronal nuclei, and tracing and measuring neurite length. The software can be downloaded at:http://bit.ly/1dQaZXa.
Image classification
Once image features are extracted, they must be classified and, here again, a number of freely available classifiers that use different classification techniques such as weighted neighborhood algorithms, gentle boosting and support vector machines (SVMs) are available to perform this task.
WND-CHARM developed by Dr. Ilya Goldberg, and his colleagues at the National Institute on Aging (Baltimore, MD;www.grc.nia.nih.gov), for example, is an image classifier that extracts high contrast features, pixel statistics, textures, and transforms of the image and filters and weights these depending on their effectiveness in discriminating between a set of previously trained images. These features are then used to classify the test images based on their similarity to the training classes using a weighted neighborhood distance algorithm. The software can be downloaded at: https://github.com/wnd-charm/wnd-charm.
Gentle boosting are two alternative classification techniques that can be found in CellProfiler. While CellProfiler, combines multiple weak classifiers to produce a significantly better strong classifier in a technique known as gentle boosting, SVM support for the software is also offered in Enhanced CellClassifier, a software package developed at ETH Zurich (Zurich, Switzerland;www.ethz.ch) by Dr. Benjamin Misselwitz.
Enhanced CellClassifier uses images analyzed by CellProfiler, and then performs multi-class classification using the SVM. The software can be downloaded at:http://bit.ly/OfhTJ0.
View other articles from our July 23D imaging e-newsletter.
Companies mentioned
Broad Institute
Cambridge, MA, USA
www.broadinstitute.org
ETH Zurich
Zurich, Switzerland
www.ethz.ch
Harvard Medical School
Boston, MA, USA
www.hms.harvard.edu
MathWorks
Natick, MA, USA
www.mathworks.com
Media Cybernetics
Rockville, MD, USA
www.mediacy.com
McGill University's Neurological Institute
Montreal, Quebec, Canada;
http://fournierlab.mcgill.ca
Molecular Devices
Sunnyvale, CA, USA
www.moleculardevices.com
National Institute on Aging
Baltimore, MD, USA
www.grc.nia.nih.gov
National Institute of Mental Health
Bethesda, MD, USA
www.nimh.nih.gov
University Medical Center Rotterdam
Rotterdam, The Netherlands
www.bigr.nl
Vision Systems Articles Archives
About the Author

Andy Wilson
Founding Editor
Founding editor of Vision Systems Design. Industry authority and author of thousands of technical articles on image processing, machine vision, and computer science.
B.Sc., Warwick University
Tel: 603-891-9115
Fax: 603-891-9297