Medical Imaging: Polarization subtraction system characterizes cancer
Breast cancer is the most common cause of death among women with over 300,000 cases being diagnosed in the US each year. To diagnose this cancer, sectioned samples of a potentially diseased tissue are specially treated and then placed onto a glass slide and examined under a microscope in a process called histopathology. Should the tissue found to be cancerous, the tumor must then be surgically removed.
"During this surgery," says Dr. Shabbir Bambot, Co-founder and CTO of Lumamed (Norcross, GA; www.lumamed.com), "it is important that the entire tumor is removed so that no cancerous tissue remains after the procedure." Because the surgeon cannot know whether any cancer remains without waiting days for histopathology, such surgical procedures may need to be repeated. This is time consuming, delays radiation treatment and the scar tissue from the original surgery makes subsequent surgery difficult.
To reduce the chances of having cancer remain, surgeons may often remove more tissue than required. Although this may reduce the number of procedures that need to be performed, more tissue may be removed than necessary.
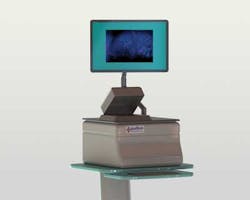
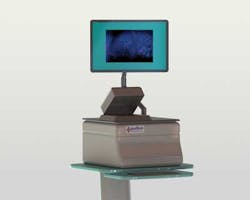
To overcome these limitations, Lumamed has developed a table top polarization microscope known as LumaScan that, following tumor excision, is used to image the tumor's exterior for signs of surface cancers.
"The LumaScan images can help the surgeon or pathologist quickly make a better decision as to whether all the cancer has been removed. Should any cancerous tissues remain, additional tissue can be removed as needed during the same surgical procedure," says Bambot. Based on Polarization Subtraction Imaging (PSI), the LumaScan system displays digital images of the surface of tissue removed during cancer.
To perform this analysis, a tissue sample is first placed onto the unit's glass platen. Light from a broadband lamp then passes through a polarizer that is mounted in the optical path of the system. This polarized light is then reflected from the tissue sample and passes through a second polarizer that is parallel to the illumination polarizer. Images from this polarizer are then captured by a CCD-based GigE camera from PixeLink (Ottawa, Ontario, Canada; www.pixelink.com) and transferred to an embedded computer with a NVIDIA GPU. A microcontroller controls the filter wheels and camera triggering.
A second polarizer rotated at 90 degrees from the first is also placed in the reflected light's optical path and a second GigE camera used to capture and transfer these images to the host computer.
"While images from the first camera will return light from the superficial layers of tissue and 50% of the randomly polarized light from deeper layers, images from the second camera with its polarizer perpendicular to the illumination polarizer will only reveal the other 50% of the randomly polarized light from the deep tissue," says Bambot. "By subtracting one image from the other, only light from the superficial surface layer will be visualized thus effectively optically sectioning tissue layer," he says.
This optical sectioning can be tailored depending on the depth of tissue to be sectioned. By placing a filter wheel in the illumination path of the system, different filters can be selected. Because longer wavelengths will penetrate the tissue sample farther than shorter wavelengths, stepping though these different wavelengths provides the surgeon with a number of images that each provide a different optical section of the tissue.
Because image subtraction must be performed in real-time as the surgeon moves the platen containing the tissue sample across the field of view of the imaging system, this subtraction must be performed rapidly.
To accomplish this task, LumaMed called upon Artemis Vision (Denver, CO, USA; www.artemisvision.com) to develop a set of image processing routines that leveraged the power of a NVIDIA GPU embedded in the device. "By transferring images from host memory to the on-board RAM of the GPU, we can offload image registration and image subtraction tasks to this processor." says Tom Brennan, President of Artemis, "Implemented this way, image registration and image subtraction tasks can be performed at live video frame rates. Thus, as the surgeon moves the platen, optically sectioned images are displayed in real-time."
LumaMed plans to develop a hand held version of the LumaScan system for dermatologists. "750,000 skin cancer surgeries a year are performed to remove complex or large cancers in a slow and painstaking process involving frozen section pathology. We believe our device could shorten this procedure by as much as two hours", says Bambot
Vision Systems Articles Archives